Welcome to the Python using Jupyter Tutorial!¶
(Python Version 2.7, 07/05/2016)
This tutorial is aimed at people who wish to use python with jupyter. If working on a computer that does not have python or jupyter installed, read and start with What you Need to get Started and Installing and Loading Python.
The best way to use this tutorial is to download the jupyter notebook and enter code and comments as you progress.
Table of Contents¶
-
What You Need to get Started
-
Installing and Loading Python
Beginning a Jupyter Session
Basic Python Syntax
Basic Python Types
Opening and Reading Files
-
Defining a Function
Creating a Class
Basic Plotting
Intermediate Plotting
Using Jupyter's Markdown Cells to Document Code
Basic Fitting
Analysis of a Fit
-
Basic Instrument Control
-
More Information
</ol>
- A computer running one of the following Operating Systems (OS): Windows 7 or greater, Mac OSX, or a unix flavor from the last 5 years.
- An internet connection.
- Some time and patience. It takes some effort to learn a new language.
# If using jupyter right now, put the cursor inside of this box and press shift-enter to see how it works
import matplotlib.pyplot as plt
# Make a data set
x=[i for i in range(-100,100)]
y=[j**2 for j in x]
# Basic Plot
plt.plot(x,y)
plt.show()
To install python, it can be downloaded from its website. However, this tutorial will use the distribution package, Anaconda.
Anaconda comes with more tools than this tutorial has time to explore. A key element of it is that it comes with the package installer __pip__. In a command line type `pip install django` and django is magically installed for the default version of python. This tutorial uses Python 2 due to it's compatibility with many of the current scientific based software packages. Python 3 is available, but many scientific packages have yet to upgrade. Once Anaconda is installed, call python from a command prompt by typing python or start a code editing tool (such as Jupyter or [PyCharm][pycharm]).
For this tutorial, you will need to download and install Anaconda for Python 2.7 (64bit version).
To begin a jupyter notebook session, open a command prompt or terminal and type jupyter notebook. This starts the server and then opens the web address http://localhost:8888/tree in the default web browser. Then click an existing notebook or open a new one.
Beginning a Jupyter Session in Windows¶
- To start a command prompt in windows
- Once in the command prompt type: jupyter notebook
- After entering the command line, the jupyter browser should automatically open the default browser at http://localhost:8888/tree.
- Click on open a new notebook
Beginning a Jupyter Session in OSX¶
- To open a terminal in OSX:
a) Click on the magnifying glass in the screen's top right corner to do a Spotlight Search
b) Type terminal in the Spotlight Search
- Once in the terminal type: jupyter notebook

- After entering the command line, the jupyter browser should automatically open the default browser at http://localhost:8889/tree.
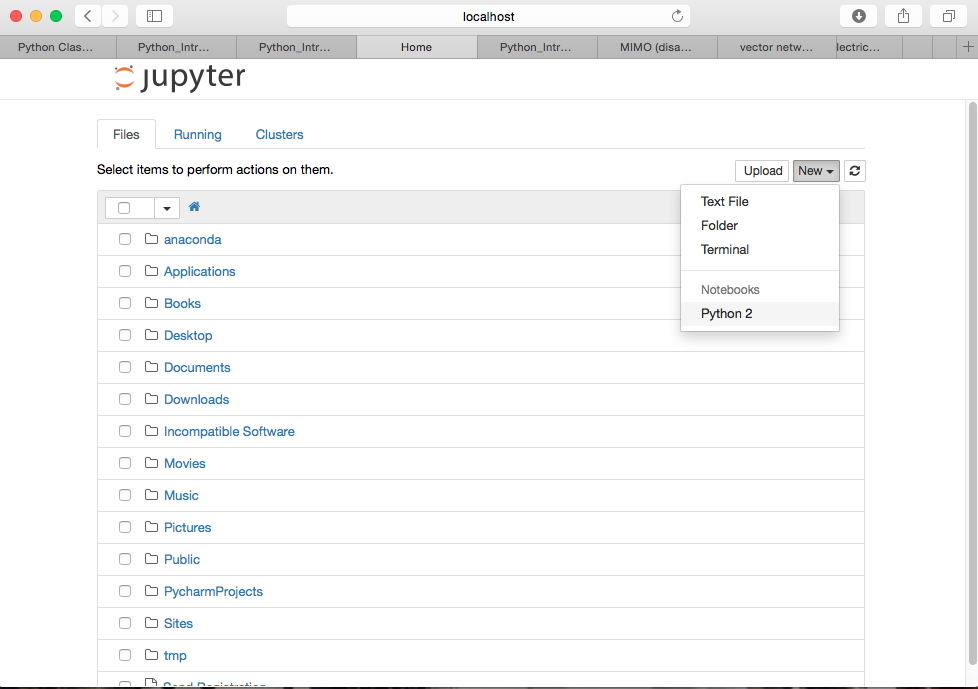
- Click on New and then Python 2 to open a new notebook.
Here are the basics of python syntax, including a few things to note for people that have used other languages:
- Whitespace is important! Every different level of code block is indented
- Flow commands and definitions of functions and classes have a colon (
:) at the end - Reserved parameters are denoted with double underscores before and after
__as in__doc__. - Comments begin with
# - Everything is zero indexed
- Power is
**not^ - No declaring variable types
Here are some examples, try them in a code cell:
# my python code
def my_function(variable_one):
"""This is a proper function definition"""
print("My function has executed and the value of variable_one is {0}".format(variable_one))
# this is an if statement
value=True
if value:
print("True!")
else:
print("False!")
# My python code that has an error
def my_function(variable_one):
"""This is an improper function definition"""
print("My function has executed and the value of variable_one is {0}".format(variable_one))
my_function('Hello There!')
# My python code
def my_function(variable_one):
"""This is a proper function definition"""
print("My function has executed and the value of variable_one is {0}".format(variable_one))
my_function('Hello There!')
# The documentation string is stored in my_function.__doc__
print(my_function.__doc__)
# This is an 'if' statement
value=True
if value:
print("True!")
else:
print("False!")
value=1
if value==1:# equality test is == not =!
print("Value is 1")
elif value<0:
print("Value is negative")
# Lists always start at index = 0
test_list_1=['a','b','c']
print("The first element of test_list_1 is {0}".format(test_list_1[0]))
test_list_2=[i for i in range(100)]
print("The first element of test_list_2 is {0}".format(test_list_2[0]))
# This is how to square a number
print(10**2)
# To square every number in a list
test_list=[i for i in range(10)]
test_list_squared=[x**2 for x in test_list]
# Text formatting for the output
print("-"*80)
print("test_list is : {0}".format(test_list))
print("-"*80)
print("test_list_squared is : {0}".format(test_list_squared))
print("-"*80)
# As a side note the ^ operator is bitwise exclusive or for ints only.
int_1=1
int_2=0
int_1^int_2
Python has some basic "types". Below are some most commonly used types.¶
- Integer is denoted by int
- String is denoted by str
- Floating point number is denoted by float
- Complex number is denoted by complex()
- List type, a list of objects that can be amended, is denoted by list
- Tuple (tuple) and set (set) are specific or special lists.
- Dictionary, an associative array, or a list with a named index are denoted by dict
# Here are some quick examples:
# Print the result of adding the numbers below
# integers
print(1+1)
print(type(1+1))
# strings
print('1'+'1')
print(type('1'+'1'))
# floats
print(1.0+1.0)
print(type(1.0+1.0))
# complex
print(1.0+1.0j)
print(type(1.0+1.0j))
# list
print([1.0]+['1'])
print(type([1.0]+['1']))
# To make lists and dictionaries
# Explicitly
my_list=[1,1,2,4,'5','space monkey']
my_dictionary={'key 1':1,'key 2':2,'key 3':my_list}
print(my_list)
print(my_dictionary)
# Add to the end of a list with append
my_list.append('New One')
print(my_list)
# Add to a dictionary by assigning the value directly
my_dictionary['New Key']='new value'
print(my_dictionary)
# Slicing lists
# To get elements 0-2
print("The first two elements of my_list are "+str(my_list[0:2]))
# or
my_list[:2]
# Last 2 elements are
my_list[-2:]
# To get the last element
# use -1 for selector
print("The last element of my_list is "+str(my_list[-1]))
One of the most basic operations for a scientist is to read in data from a file and prepare it for analysis. There are many libraries that exist for python which have many of the difficult parts of the code already complete. In an effort to better understand python, this tutorial will cover the "manual" way to read in the data vs using a library. In particular, this will cover some of the functionality in numpy.loadtxt() and panadas.read_csv(). It is important to note that the libraries of choice would be numpy or pandas.
# # THIS IS THE CODE TO WRITE THE EXAMPLE FILE IN CASE IT DOES NOT ALREADY EXIST.
# # IT PROBABLY WILL NOT WORK UNLESS PYMEASURE HAS BEEN INSTALLED PROPERLY.
# # A detour to write the needed example file:
# import os
# import scipy
# import numpy as np
# import random
# from pyMez import *
# start=-10
# stop=10
# num=1000
# xdata=np.linspace(start,stop,num)
# ydata_with_noise_2=[.5*scipy.exp(-(x-.1)**2/(2.0*3.0**2))+random.gauss(0,.03) for x in xdata]
# # This gives the ability to look for files
# #os.listdir(os.getcwd()) #list all the files in the current working directory
# # This should be the directory where the example files are located.
# file_directory=os.path.join(os.getcwd(),'Python_Introduction_Files')
# # If the files are in the right place, open the following:
# file_name='Example_File_1.txt'
# data=[]
# for index,item in enumerate(xdata.tolist()):
# data.append([item,ydata_with_noise_2[index]])
# options={"data":data,"column_names":['Position','Amplitude'],"column_types":['float','float']}
# table=AsciiDataTable(None,**options)
# table.path=os.path.join(file_directory,file_name)
# table.save()
# To open a file
# First it is useful to ‘import os’, a library that deals with the operating system.
import os
# This gives the ability to look for files
#os.listdir(os.getcwd()) #list all the files in the current working directory
# This should be the directory where the example files are located.
file_directory=os.path.join(os.getcwd(),'Python_Introduction_Files')
# If the files are in the right place, open the following:
file_name='Example_File_1.txt'
file_path=os.path.join(file_directory,file_name)
# To open the file use the built in function open, in the read mode.
in_file=open(file_path,'r')
# The complete contents of the file can be retrieved by use
# of ‘contents=in_file.read()’, however it is preferable for processing
# reasons, to use a pattern that reads each line of the file into a
# separate element of a list.
lines=[]
for line in in_file:
lines.append(line)
# The list named lines contains the information in the file. It is a list of strings.
print("After reading lines[0] and lines[1] are now {0}".format(lines[0:2]))
# It is good practice to close in_file
in_file.close()
# It is necessary at this point to conver the list of strings to a list of numbers.
# The conversion requires the use of float() and split.
# First convert each line into a list instead of a single string. This is accomplished via the split() method.
# The first line of the file contains a column heading so the conversion process should begin with the
# Second line.
lines=[line.split(',') for line in lines[1:]]
print("After splitting lines[0] is now {0}".format(lines[0]))
# Next convert all values to the type 'float' instead of 'str'
lines=[map(lambda x:float(x.rstrip().lstrip()),row) for row in lines]
print("After converting lines[0] is now {0}".format(lines[0]))
A function can have none, one, or many arguments. A function can also return a value or not. This section will cover how to create a function that does what the code in the previous section did.
First, a few important things to remember about functions:
- Functions start with the keyword
def - The name of a function cannot have any spaces.
- The name of a function cannot start with a number.
- Functions end when the indented block ends.
Below is a basic example to show how it works:
def function_one(variable_one,variable_two='default value',*list_arguments,**keyword_arguments):
"""function_one is an example function that will take a positionial argument, a named argument or the "packed" arguments
in list_arguments or keyword arguments, it prints them out but does not return a value"""
print("{0}, is the value of variable_one and {1} is the value of variable_two".format(variable_one,variable_two))
for index,item in enumerate(list_arguments):
print("Item number {0} in list_arguments is {1}".format(index,item))
for key, value in keyword_arguments:
print("keyword_arguments[{0}]={1}".format(key,value))
def function_one(variable_one,variable_two='default value',*list_arguments,**keyword_arguments):
"""function_one is an example function that will take a positionial argument, a named argument or the "packed" arguments
in list_arguments or keyword arguments, it prints them out but does not return a value"""
print("{0}, is the value of variable_one and {1} is the value of variable_two".format(variable_one,variable_two))
for index,item in enumerate(list_arguments):
print("Item number {0} in list_arguments is {1}".format(index,item))
for key, value in keyword_arguments.items():
print("keyword_arguments[{0}]={1}".format(key,value))
function_one('Hello',"hi",*['a list','of','values','or',123],**{'A Key':'A Value'})
function_one('Hello again',*['a list','of','values','or',123],**{'A Key':'A Value'})
function_one(*['a list','of','values','or',123],**{'A Key':'A Value'})
function_one(**{'A Key':'A Value','variable_one':1})
# The code below uses a function to do the same thing as the code in the 'opening a file section'.
# Note: Do not forget the doc strings.
def process_data_file(file_path,skiprows=1):
"""process_data_file is a function that opens a data file and returns a 2-d list"""
in_file=open(file_path,'r')
x_data=[]
y_data=[]
for index,line in enumerate(in_file):
if index>skiprows-1:
line_list=line.split(',')
line_list=list(map(lambda x:float(x),line_list))
x_data.append(line_list[0])
y_data.append(line_list[1])
elif index<=skiprows-1:
pass
return [x_data,y_data]
# Try the function
#process_data_file(os.path.join(file_directory,file_name))
A class is a block of code, including variables and functions, that lives on its own. By defining chunks of code with classes it is stating: "these are important functions and variables when dealing with this problem". It is impossible to teach in a limited amount of time how functional, procedural, and object oriented programming (OOP) methodologies differ. OOP can be extremely helpful and/or necessary when programming for scientific purposes. When dealing with the repeated opening and analyzing of the same kind of file, the code and useful functions that do this should live in one or a few classes. Another time when it is helpful to know OOP is when dealing with any Graphical User Interface (GUI). Most GUI toolboxes use classes.
class CSVData():
"""CSVData is a class that holds example data"""
def __init__(self,file_path=None,**options):
"""Intializes the CSVData class, requires a file_path"""
# This pattern allows for the setting of defaults for a large number of options
# It also provides the ability to potentially add options after the initial class is created
defaults={'directory':None,'file_name':None,'skiprows':1}
self.options={}
for key,value in defaults.items():
self.options[key]=value
for key,value in options.items():
self.options[key]=value
# Using the function from above and some error handling.
# If there is an error in the code inside of the try: block it will be sent to an except block
try:
# First see if the user set file_path or directory and file_name in options
if file_path is None:
file_path=os.path.join(self.options['directory'],self.options["file_name"])
# Then use the function process_data_file to make the x_data and y_data attributes
[self.x_data,self.y_data]=process_data_file(file_path)
except:
print("There was an error in opening file {0}".format(file_path))
raise
# Now use the class on the original file
new_data=CSVData(os.path.join(os.getcwd(),'Python_Introduction_Files','Example_File_1.txt'))
#print new_data.x_data
Those steps might seem pointless, but were preparation for the fact that through functions and variables, those chunks of code can be passed on to make more complicated programs. This is called class inheritance.
# Example of an inherited class
class MyData(CSVData):
"""MyData is a class that opens a standard CSV file and keeps things important to me """
def __init__(self,file_name,**options):
defaults={'directory':None,'file_name':None,'skiprows':1}
self.options={}
for key,value in defaults.items():
self.options[key]=value
for key,value in options.items():
self.options[key]=value
CSVData.__init__(self,file_name,**self.options)
self.max=max(self.y_data)
self.min=min(self.y_data)
new_data=MyData(os.path.join(os.getcwd(),'Python_Introduction_Files','Example_File_1.txt'))
new_data.max
Now the class MyData has all of the functions and variables in CSVData in addition to the new ones!!
Basic plots that emulate early MATLAB plots are easy to make in python using the library matplotlib.
# Basic plotting with Matplotlib
import matplotlib.pyplot as plt
# Make a data set
x=[i for i in range(-100,100)]
y=[j**2 for j in x]
# Basic Plot
plt.plot(x,y)
plt.show()
If making a publication quality plot, many changes to suit the given audience are necessary such as font size, markers, etc. In addition, it would be nice to have the plot inside the notebook for reference; this is achieved with the notebook "magic". There are a lot of functionalities hidden in jupyter (and its predecessor ipython) that can be accessed using magics. A magic is started by putting something like %matplotlib notebook at the beginning of a cell. In this case it will cause the plot to live in the notebook and be interactive.
Making an Interactive Plot¶
Note: To undo an interactive plot usually requires a kernel restart.
%matplotlib notebook
x=[i for i in range(-100,100)]
y=[j**2 for j in x]
plt.plot(x,y)
plt.show()
# The magic stays until the kernel is restarted, so now is a good opportunity to add a plot_title
plt.title('Parabolic')
plt.show()
# This will spruce up the title with a little latex
plt.title('Parabolic Curve '+r'$f(x)=x^2$')
# Adding x and y labels
plt.xlabel('X Value')
plt.ylabel('Y Value')
# Changing the line width
line=plt.plot(x,y)
plt.setp(line,color='r', linewidth=2.0)
# Switch from a line to markers
plt.plot(x,y,'ro')
# Changing the fontsize
plt.xlabel('X Value',fontsize=24)
# Adding annotations in the coordinates system of the plot
x,y=-50,7000
plt.text(x, y, 'A New text Annotation at({0:3.2f},{1:3.2f})'.format(x,y))
# To kill the current plot
plt.close()
The jupyter environment comes with a handy way of creating notes and documenting code - the markdown cell. A markdown cell handles traditional markdown (a HTML short hand), most html tags, and latex. This allows the user to make their code pretty.
In this section the user will learn:
- How to create simple markdown cells with formatted text inside
- How to add html for more complex formatting
- How to highlight code for demonstration, but not execution
- How to output simple formulae using latex (note: sometimes this is broken in Internet Explorer, use another browser such as Chrome)
Creating a Simple Markdown cell with Formatted Text Inside¶
- Create a new cell, by executing the previous cell or Insert Cell
- Change the type of cell from Code to Markdown by escape+m or by changing the selection in the tool bar
- In the new cell enter
# My Heading, execute the cell by pressing shift+enter. This will produce something like: # My Heading - The markdown shorthand has the ability to apply a large number of simple styles such as bold, italic, tables, links, and the equivalent of the HTML heading tags. A more complete listing of the possibilities for markdown can be found at :https://daringfireball.net/projects/markdown/syntax a more jupyter focused tutorial can be found here.
Most used shorthand rules:
- Headings start with
#. The level is given by the number of#'s, like## Heading 2# Heading 1 ## Heading 2 ### Heading 3 #### Heading 4 - Number lists are default for things that start with numbers like
1. Numbered list element 1- Numbered list element 1
- Numbered list element 2
- Unordered or bulleted lists start with a symbol, such as
+
+ Bullet 1- Bullet 1
- Bullet 2
- Bold is two underscores before and after the word or phrase, like
__my bolded text__and will be displayed as my bolded text - Italics is an asterix before and after, like
*italics is awesome*and will be displayed as italics is awesome - Tables can be made with underscores and pipes such as
| Tables | Are | Cool || Tables | Are | Cool |, I usually use the html version though.
Adding HTML for more Complex Formatting¶
In addition to handling the markdown shorthand syntax, jupyter's markdown cells handle most HTML tags properly. These include the ability to add the style attribute.
For example, to change a heading to be red and in a different font type <h1 style="color:red;font-family:serif"> My Heading</h1>, execute the cell and something similar to the heading below should appear.
My Heading
or to do a styled table such as:
<table style="color:blue;"><tr><td style="border: 5px solid green;">Table row 1 column 1 </td><td style="border: 5px solid blue;">Table row 1 column 2 </td></tr></table>
| Table row 1 column 1 | Table row 1 column 2 |
this works for a range of tags that include :
<h1></h1>...<h6></h6><p></p><table></table><img />
does not work for javascript or svg. To embed them requires the use of the rich output methods in jupyter.
Code Highlighting for Code not Executed¶
Markdown cells have the ability to include a chunk of code in number of languages, provided they are properly identified with highlighting. To do this, start by escaping the test with three ` (backquote, the character under ~ on most keyboards) followed by a language name. So it looks like ```python ```
For example:
```python
# python comment
```
is rendered like
# python comment
This works with HTML, javscript, python and a few others by changing the word directly after the three backquotes.
HTML:
<h3 style="color:red"> Table </h3>
<table> </table>
Javascript:
// A javascript comment
function myJavascriptFunction(input){
var longUnreadableName=null;
document.getElementById('fake-id').innerHTML=input;
}
Visual Basic:
vb
dim Variable as integerLatex for Equations and Type Setting¶
Jupyter markdown cells also understand latex. To do this, wrap expressions with two \$'s
i.e.:
```$$your latex here$$``` for equations on their own line
```$your latex here$``` for inline equations
This is sometimes broken in Internet Explorer, but works fine in most other browsers.
For example:
the sum of xi from i=0 to i=100
`$\sum\limits{i=100}^{0}x{i}$`
renders as:
$\sum\limits_{i=100}^{0}x_i$
or a gaussian is `$$f(x)=ae^{-{(x-x_0)}^2\over{2\sigma^2}}$$`:
$$f(x)=ae^{-{(x-x_0)}^2\over{2\sigma^2}}$$
Fitting Using scipy¶
There is more than one way to do a fit in python. This tutorial will use the scipy library and its optimizing package to do a fit. Scipy can be found at https://www.scipy.org/
# Import scipy and some of its important modules
import scipy
import scipy.optimize
import scipy.stats
# Import numpy for linspace and random numbers
import numpy as np
# Import python random numbers
import random
# Simulate data for fit
start=-10
stop=10
num=1000
xdata=np.linspace(start,stop,num)
ydata=.5*scipy.exp(-(xdata-.1)**2/(2.0*3.0**2))
ydata_with_noise=[.5*scipy.exp(-(x-.1)**2/(2.0*3.0**2))+np.random.random() for x in xdata]
ydata_with_noise_2=[.5*scipy.exp(-(x-.1)**2/(2.0*3.0**2))+random.gauss(0,.03) for x in xdata]
# Define fitting functions
def lorentzian_function(a,x):
"a[0]=amplitude,a[1]=center,a[2]=FWHM"
return a[0]*1/(1+(x-a[1])**2/(a[2]**2))
def gaussian_function(a,x):
" a[0]=amplitude, a[1]=center, a[2]=std deviation"
return a[0]*scipy.exp(-(x-a[1])**2/(2.0*a[2]**2))
# Define the fit function that returns least square optimization
def fit(function,xdata,ydata,a0):
fit_dictionary={}
error_function=lambda a, xdata, ydata:function(a,xdata)-ydata
a,succes=scipy.optimize.leastsq(error_function, a0,args=(xdata,ydata))
return a
# Make a horrible guess
a0=[1,1,1]
# Call fit function on the data
result=fit(gaussian_function,xdata,ydata_with_noise_2,a0)
#result=fit(lorentzian_function,xdata,ydata_with_noise_2,a0)
# Plot the results
# Data plot
plt.plot(xdata,ydata_with_noise_2,'bo')
# Fit plot
plt.plot(xdata,[gaussian_function(result,x) for x in xdata],'r-',linewidth=3)
# Some plot type setting
plt.title('Fit of data')
options={'fontsize':24}
# Text showing the numeric values with latex
plt.text(-8, .5, r'$%3.2fe^{\frac{-{(x-%3.2f)}^2}{2*%3.2f^2}}$'%(result[0],result[1],result[2]),**options)
plt.show()
First Calculate the Residuals¶
Probably the most basic analysis of a fit is that of computing how the fit varies from the data. These data points are typically called residuals.
plt.close()
def calculate_residuals(fit_function,a,xdata,ydata):
"""Given the fit function, a parameter vector, xdata, and ydata returns the residuals as [x,y] pairs"""
output_x=xdata
output_y=[fit_function(a,x)-ydata[index] for index,x in enumerate(xdata)]
return [output_x,output_y]
[x,y]=calculate_residuals(gaussian_function,result,xdata,ydata_with_noise_2)
plt.plot(x,y)
#calculate_residuals(gaussian_function,result,xdata,ydata_with_noise_2)
Look at the distribution of residuals. How does the data vary from the model (fit)?¶
# To do a histogram of the residuals
plt.close()
plt.hist(y)
# To find the mean of the residuals and the standard deviation
import numpy as np
mean_y=np.mean(y)
std_y=np.std(y)
print("The residuals mean is {0} \n The residuals standard deviation is {1}".format(mean_y,std_y))
It is Useful to Look at all the Information at the Same Time¶
plt.close()
# It is often helpful to see all of the above together at the same time
def fit_and_plot(fit_function,xdata,ydata,a0=[1,1,1]):
"""fit and plot, fits xdata, ydata with fit function and returns a plot of the fit overlaid with the data and
a plot of the residuals, including a histogram"""
fit_results=fit(fit_function,xdata,ydata,a0)
# To do 2 plots in 2 rows with shared axis
fig, (ax0, ax1) = plt.subplots(nrows=2, sharex=True)
ax0.plot(xdata,ydata,'bo',label='data')
ax0.plot(xdata,[fit_function(fit_results,x) for x in xdata],'r-',linewidth=3,label='fit')
ax0.legend(loc='upper right', shadow=True)
ax0.set_title('Fit of data using {0}'.format(fit_function.__name__))
[x,y]=calculate_residuals(fit_function,fit_results,xdata,ydata)
ax1.plot(x,y)
ax1.set_title('Residuals')
plt.show()
fit_and_plot(gaussian_function,xdata,ydata_with_noise_2)
When the data does not Conform to the Model it is Common to see Structured Residuals¶
plt.close()
# Try the same data with a different fit function
fit_and_plot(lorentzian_function,xdata,ydata_with_noise_2)
plt.close()
a0=[1 for i in range(3)]
[x,y]=calculate_residuals(lorentzian_function,fit(lorentzian_function,
xdata,ydata_with_noise_2,a0),xdata,ydata_with_noise_2)
plt.hist(y)
plt.show()
mean_y=np.mean(y)
std_y=np.std(y)
print("The residuals mean is {0} \nThe residuals standard deviation is {1}".format(mean_y,std_y))
%matplotlib inline
from ipywidgets import *
import numpy,scipy,random
t=numpy.arange(0.0,1.0,0.01)
start=-10
stop=10
num=1000
xdata=numpy.linspace(start,stop,num)
ydata=.5*scipy.exp(-(xdata-.1)**2/(2.0*3.0**2))
ydata_with_noise=[.5*scipy.exp(-(x-.1)**2/(2.0*3.0**2))+numpy.random.random() for x in xdata]
ydata_with_noise_2=[.5*scipy.exp(-(x-.1)**2/(2.0*3.0**2))+random.gauss(0,.03) for x in xdata]
interact(lambda noise_sigma:fit_and_plot(lorentzian_function,
xdata,
[.5*scipy.exp(-(x-.1)**2/(2.0*3.0**2))+random.gauss(0,noise_sigma) for x in xdata]),
noise_sigma=(0,.3,.01))
interact(lambda noise_sigma,noise_mu:fit_and_plot(gaussian_function,
xdata,
[.5*scipy.exp(-(x-.1)**2/(2.0*3.0**2))+random.gauss(noise_mu,noise_sigma) for x in xdata]),
noise_sigma=(0,1,.01),noise_mu=(0,1,.01))
Visa instrument control requires a visa driver for your OS. The standard one is made by National Instruments, but there are others.¶
- Install NIVisa or on Mac OS https://pyvisa.readthedocs.io/en/stable/getting_nivisa.html#getting-nivisa
- Install pyvisa (from the OS command prompt type
pip install pyvisaor from jupyter try:import os os.system('pip install pyvisa')
# Test the installation
import visa
rm = visa.ResourceManager()
print(rm.list_resources())
# To send a gpib command, change the address to the device
my_instrument = rm.open_resource('GPIB0::14::INSTR')
print(my_instrument.query('*IDN?'))
Other Resources¶
This tutorial only: https://pyvisa.readthedocs.io/en/stable/getting_nivisa.html#getting-nivisa covers a fraction of available resources for python, for more information see the links below.
Python Based Packages Like Anaconda¶
- Enthought https://www.enthought.com/products/canopy/
- Python(x,y)https://python-xy.github.io/